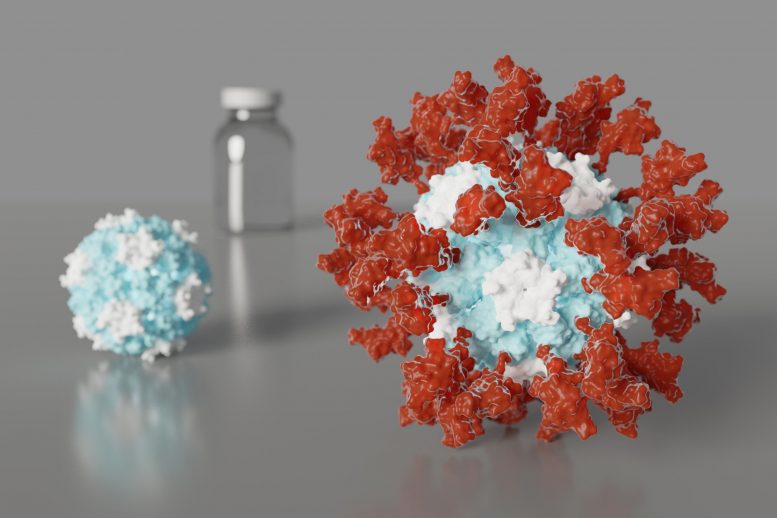
Artist’s depiction of an ultrapotent COVID-19 vaccine candidate in which 60 pieces of a coronavirus protein (red) decorate nanoparticles (blue and white). The vaccine candidate was designed using methods developed at the UW Medicine Institute for Protein Design. The molecular structure of the vaccine roughly mimics that of a virus, which may account for its enhanced ability to provoke an immune response. Credit: Ian Haydon/ UW Medicine Institute for Protein Design
Preclinical data published in Cell show the nanoparticle vaccine spurs extremely high levels of protective antibodies in animal models.
An innovative nanoparticle vaccine candidate for the pandemic coronavirus produces virus-neutralizing antibodies in mice at levels 10 times greater than is seen in people who have recovered from COVID-19 infections. Designed by scientists at the University of Washington School of Medicine in Seattle, the vaccine candidate has been transferred to two companies for clinical development.
Compared to vaccination with the soluble SARS-CoV-2 Spike protein, on which many leading COVID-19 vaccine candidates are based, the new nanoparticle vaccine produced 10 times more neutralizing antibodies in mice, even at a sixfold lower dose. The data also show a strong B-cell response after immunization, which can be critical for immune memory and a durable vaccine effect. When administered to a single nonhuman primate, the nanoparticle vaccine produced neutralizing antibodies targeting multiple different sites on the Spike protein. Researchers say this may ensure protection against mutated strains of the virus, should they arise. The Spike protein is part of the coronavirus infectivity machinery.
The findings were published on October 30, 2020, in Cell. The lead authors of this paper are Alexandra Walls, a research scientist in the laboratory of David Veesler, a UW associate professor of biochemistry; and Brooke Fiala, a research scientist in the lab of Neil King, UW assistant professor of biochemistry.
The vaccine candidate was developed using structure-based vaccine design techniques invented at UW Medicine. It is a self-assembling protein nanoparticle that displays 60 copies of the SARS-CoV-2 Spike protein’s receptor-binding domain in a highly immunogenic array. The molecular structure of the vaccine roughly mimics that of a virus, which may account for its enhanced ability to provoke an immune response.
“We hope that our nanoparticle platform may help fight this pandemic that is causing so much damage to our world,” said King, inventor of the computational vaccine design technology at the Institute for Protein Design. “The potency, stability, and manufacturability of this vaccine candidate differentiate it from many others under investigation.”
Hundreds of candidate vaccines for COVID-19 are in development around the world. Many require large doses, complex manufacturing, and cold-chain shipping and storage. An ultrapotent vaccine that is safe, effective at low doses, simple to produce and stable outside of a freezer could enable vaccination against COVID-19 on a global scale.
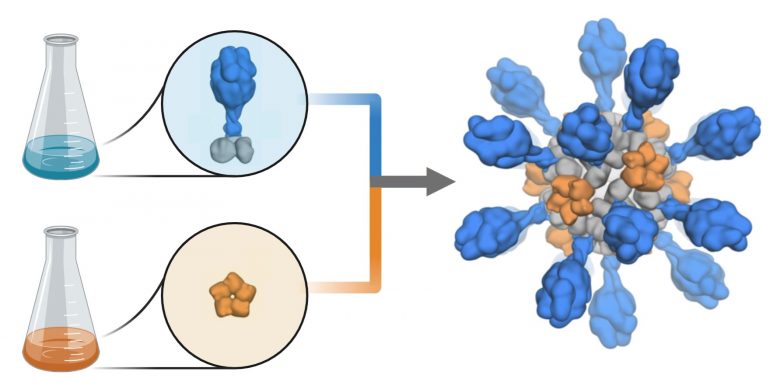
Production schematic shows how coronavirus proteins are added to a computer-designed nanoparticle platform to create a candidate vaccine against COVID-19. The vaccine candidate was designed and tested in animal models by researchers at the University of Washington School of Medicine. Credit: Ian Haydon/UW Medicine Institute for Protein Design
“I am delighted that our studies of antibody responses to coronaviruses led to the design of this promising vaccine candidate,” said Veesler, who spearheaded the concept of a multivalent, receptor-binding, domain-based vaccine.
The lead vaccine candidate from this report is being licensed non-exclusively and royalty-free during the pandemic by the University of Washington. One licensee, Icosavax, a Seattle biotechnology company co-founded in 2019 by King, is currently advancing studies to support regulatory filings and has initiated the U.S. Food and Drug Administration’s Good Manufacturing Practice. To accelerate progress by Icosavax to the clinic, Amgen has agreed to manufacture a key intermediate for these initial clinical studies. Another licensee, SK bioscience of South Korea, is advancing its own studies to support clinical and further development.
Reference: “Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2Alexandra C. Walls, Brooke Fiala, Alexandra Schäfer, Samuel Wrenn, Minh N. Pham, Michael Murphy, Longping V. Tse, Laila Shehata, Megan A. O’Connor, Chengbo Chen, Mary Jane Navarro, Marcos C. Miranda, Deleah Pettie, Rashmi Ravichandran, John C. Kraft, Cassandra Ogohara, Anne Palser, Sara Chalk, E-Chiang Lee, Kathryn Guerriero, Elizabeth Kepl, Cameron M. Chow, Claire Sydeman, Edgar A. Hodge, Brieann Brown, Jim T. Fuller, Kenneth H. Dinnon, III, Lisa E. Gralinski, Sarah R. Leist, Kendra L. Gully, Thomas B. Lewis, Miklos Guttman, Helen Y. Chu, Kelly K. Lee, Deborah H. Fuller, Ralph S. Baric, Paul Kellam, Lauren Carter, Marion Pepper, Timothy P. Sheahan, David Veesler and Neil P. King, Accepted 26 October 2020, Cell.
DOI: 10.1016/j.cell.2020.10.043
The research reported in Cell was supported by the National Institute of Allergy and Infectious Diseases (DP1AI158186, HHSN272201700059C, 3U01AI42001-02S1), National Institute of General Medical Sciences (R01GM120553, R01GM099989), Bill & Melinda Gates Foundation (OPP1156262, OPP1126258, OPP1159947), Defense Threat Reduction Agency (HDTRA1-18-1-0001), Pew Biomedical Scholars Award, Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund, Fast Grants, Animal Models Contract HHSN272201700036I-75N93020F00001, University of Washington’s Proteomics Resource (UWPR95794), North Carolina Coronavirus Relief Fund, and gifts from Jodi Green and Mike Halperin and from The Audacious Project.