Researchers in the United States have developed an inhibitor of the spike protein found on the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that limits its formation in host human cells that would otherwise be the source of newly generated virions.
The SARS-CoV-2 virus is the agent responsible for the ongoing coronavirus disease 2019 (COVID-19) pandemic and the spike protein is the main structure the virus relies on for host cell entry.
Importantly, the inhibitor was effective against the spike proteins of other coronaviruses, including SARS-CoV-1 and Middle East respiratory syndrome CoV (MERS-CoV).
Furthermore, the researchers say the polypeptide inhibitor – called F1 – is expected to be effective against the spike proteins of almost any SARS-CoV-2 variants that may emerge in the future.
“We expect the inhibitor reported here to be an invaluable aid to help end the COVID-19 pandemic,” writes Jianpeng Ma and colleagues from Baylor College of Medicine in Houston, Texas.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.
.jpg)
Study: High-Potency Polypeptide-based Interference for Coronavirus Spike Glycoproteins. Image Credit: NIAID
Coronaviruses have posed a major threat for two decades
Within just the last 20 years, three coronaviruses have posed a significant threat to public health, causing regional and global outbreaks of potentially life-threatening respiratory disease.
These include the SARS-CoV-1 virus responsible for the 2002 to 2003 SARS outbreak, the MERS-CoV virus that has caused various outbreaks across the Middle East since 2012, and the novel SARS-CoV-2 virus that is responsible for the ongoing COVID-19 pandemic.
Currently, researchers are racing to develop vaccines based on the SARS-CoV-2 spike protein that will generate immune responses against the wild-type spike following natural infection with the virus.
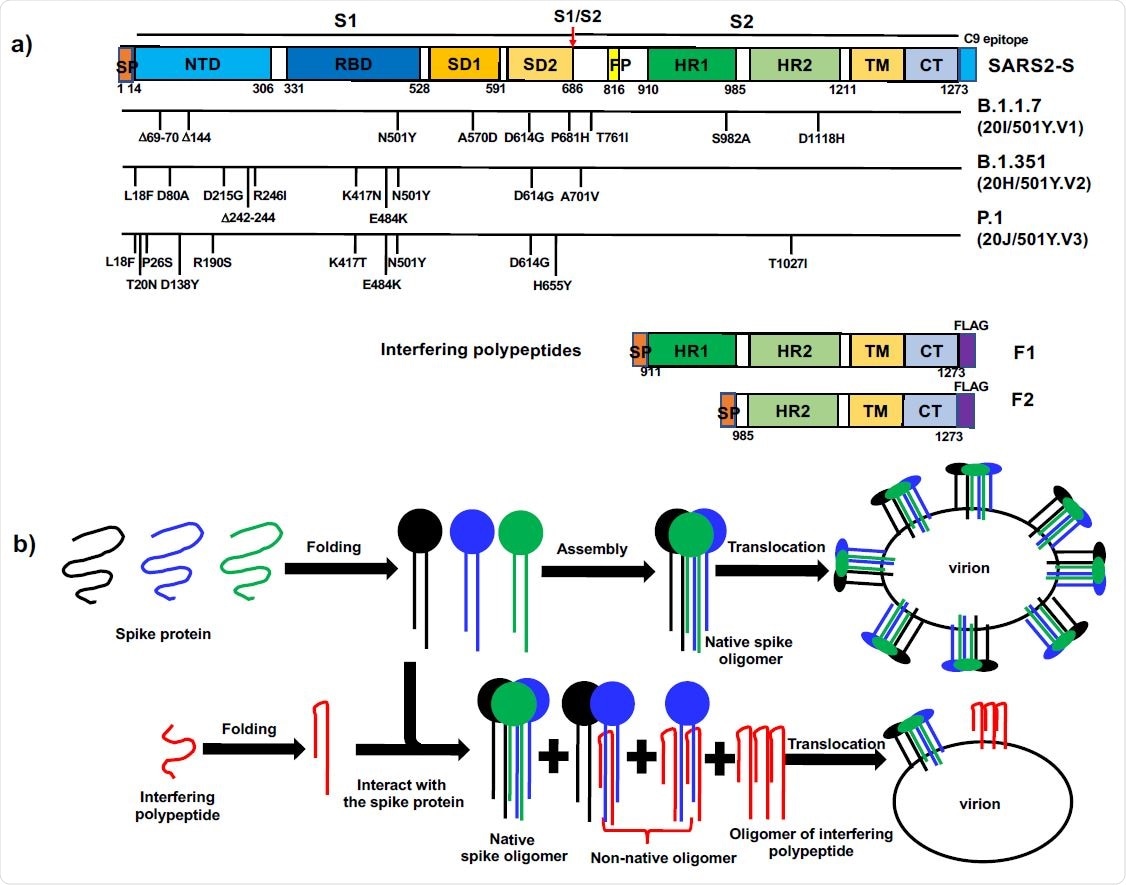
The concept of polypeptide-based protein interference against coronavirus spike proteins. a). Domain organization of COVID-19 SARS2-S, the mutations in recent variants and the design of interfering polypeptides F1 and F2. SP: Signal peptide; NTD: N-terminal domain; RBD: receptor-binding domain; SD1: subdomain 1; SD2: subdomain 2; FP: fusion peptide; HR1: heptad repeat 1; HR2: heptad repeat 2; TM: transmembrane domain; CT: Cytoplasmic tail. The cleavage at S1/S2 (red arrow) gives rise to N-terminal S1 fragment and C-terminal S2 fragment. The signal peptide sequence at the extreme N-termini of F1 and F2 allowed the polypeptides to be translocated in the same way as COVID-19 SARS2-S. At the extreme C-termini, SARS2-S had a C9 epitope recognized by C9-rhodopsin antibody 1D4, while both F1 and F2 had a FLAG-tag. b). Diagram of polypeptide-based interference targeting coronavirus spike proteins. Top row: in the normal situation, the spike proteins were synthesized, folded and formed native spike oligomers, which were anchored on virion envelope. Bottom row, interfering polypeptides formed nonnative oligomers with the wild-type spike proteins, thus reducing the level of native spike oligomers on the envelope of new virions.
The emergence of variants means new approaches are urgently needed
Once the SARS-CoV-2 spike protein binds to its host cell receptor – angiotensin-converting enzyme 2 (ACE2) – the spike is cleaved into two subunits.
Subunit 1 (S1) is the main target of neutralizing antibodies following natural infection or vaccination and is therefore under constant positive selection for immune escape variants. Subunit 2 (S2), on the other hand, is more conserved between different coronavirus strains.
Since SARS-CoV-2 was first identified in Wuhan, China, in late December 2019, its unprecedented spread has led to the emergence of several variants harboring extensive mutations in the spike protein.
Some of these variants have exhibited tighter binding to ACE2 and increased transmissibility, as well as partial resistance to antibody neutralization by sera from vaccinated or convalescent individuals.
“With over 130 million confirmed cases and widespread vaccination around the globe, the emergence of new escape SARS-CoV-2 variants could be accelerated,” says Ma and colleagues. “New therapeutics insensitive to mutations are thus urgently needed.”
The concept behind the current study
Following host cell entry, the SARS-CoV-2 genome guides the synthesis of new spike proteins. The proteins are then folded, assembled and translocated for interaction with newly replicated genomic RNA to generate new virions.
Ma and colleagues hypothesized that foldable fragments of the spike protein such as polypeptides derived from S2 would form non-native oligomers with wild-type spikes. This would reduce the level of the native spike on the envelope of newly generated virions and potentially impair their infectivity, says the team.
The researchers synthesized a polypeptide called F1 that contained part of the sequence from S2 of the SARS-CoV-2 spike protein. They then tested its impact on the expression and cell surface translocation of spike proteins to the host cell surface in the human cell line HEK293T.
What did the study find?
Transfection of the cells with SARS-CoV-2 spike-harboring plasmid yielded high expression of cleaved spike proteins in the whole cell lysate.
When the F1-harboring plasmid was co-transfected with the spike-harboring plasmid, spike S2 was almost completely diminished in the whole cell lysate and in the cell surface fraction.
“Thus, F1 strongly interfered with the expression and cell surface translocation of SARS-CoV-2 spike,” says Ma and colleagues.
Although F1 was derived from the SARS-CoV-2 sequence, the inhibitor was equally as effective against the spike proteins of SARS-CoV-1 and MERS-CoV. Again, S2 was almost completely diminished in the whole cell lysate and the cell surface fraction.
The amino acid sequence identity shared between these different coronavirus spikes was as low as 35%, suggesting that F1 could be highly resistant to mutations in the spike sequences of newly emerging SARS-CoV-2 variants.
The agent could be effective against coronaviruses over “a long time period”
“The high potency of F1 in interfering with expression and surface translocation of the spike glycoproteins from coronaviruses that caused severe outbreaks or pandemics between 2002 and 2021 suggests that F1 has a high promise to become an effective therapeutic agent against different coronavirus lineages over a long time period,” write the researchers.
Furthermore, since the sequences corresponding to the F1 polypeptide are highly conserved between SARS-CoV-2 variants, this inhibitor can be expected to be effective against the spike proteins of almost any variants that emerge in the future, they add.
“We expect the inhibitor reported here to be an invaluable aid in the effort to stop the COVID-19 pandemic,” concludes the team.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.