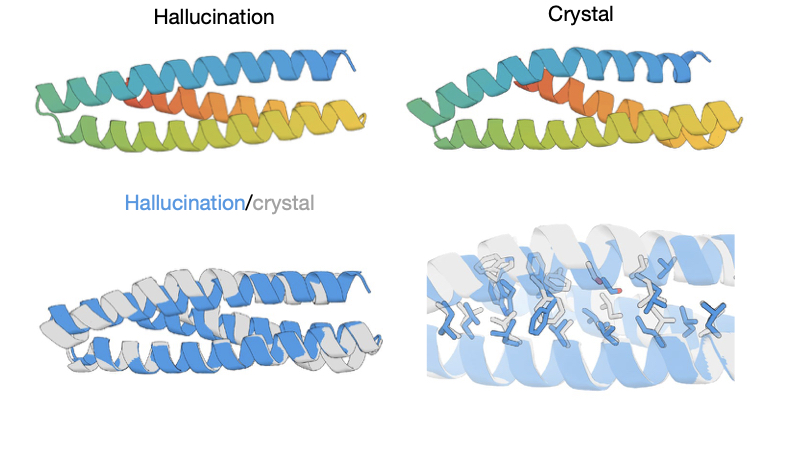
/ Top row: the hallucination and actual structure. Bottom row: the two structures superimposed.
Anishchenko et. al.
Chemically, proteins are just a long string of amino acids. Their amazing properties come about because that chain can fold up into a complex, three-dimensional shape. So understanding the rules that govern this folding could not only give us insights into the proteins that life uses but could potentially help us design new proteins with novel chemical abilities.
There’s been remarkable progress on the first half of that problem recently: researchers have tuned AIs to sort through the evolutionary relationships among proteins and relate common features to structures. As of yet, however, those algorithms aren’t any help for designing new proteins from scratch. But that may change, thanks to the methods described in a paper released on Wednesday.
In it, a large team of researchers describes what it terms protein “hallucinations.” These are the products of a process that resembles a game of hotter/colder with an algorithm, starting with a random sequence of amino acids, making a change, and asking, “Does this look more or less like a structured protein?” Several of the results were tested and do, in fact, fold up like they were predicted to.
AI hallucinations
The odd terminology here can’t be blamed on the authors of the new paper. Instead, the term “hallucination” was applied to work done by Google’s AI team. That work involved starting with an image of random pixels and asking a neural network trained to recognize fruit, “How much does this look like a banana?” After some random tweaks, the question was asked again; any changes that increased the image’s banana-like properties were retained, and the process repeated.
The end result clearly has banana-like aspects, but it looks more like a cubist and impressionist both had a go at the bananas before running a few random Photoshop filters. While the term isn’t used in Google’s blog post, others labeled the images “hallucinations.”

Random noise (left) gets converted to a banana-like hallucination (right) by repeated queries to a banana-recognition AI.
The researchers thought that, if this works for AIs that handle image recognition, maybe it would also work with AIs that suggest 3D structures for proteins.
Those of you paying careful attention here may notice a problem, however. The biology-specific algorithms don’t output a rating of whether something is structure-like; instead, they simply assume there’s a structure and try to suggest what it is. So they’re not inherently set up to do the sort of getting-hotter/getting-colder evaluation that’s needed to create a hallucination.
The research team figured out a way around this, however. Unstructured proteins tend to spread out in space, with only a handful of neighboring amino acids interacting with each other. Highly structured proteins, by contrast, tend to be compact and fold so that amino acids in different parts of the chain can interact with each other. The algorithm they were using for structure prediction, trRosetta, outputs its predictions as the relative location of each amino acid in a 3D space. So, by using a measure of their spread, the authors were able to provide a sort of answer to the question “how structured does this look?”
Starting from random
To start their structural hallucinations, the researchers generated numerous proteins composed of 100 random amino acids and fed them to the trRosetta software. As expected, all of the proteins were unstructured at the start. Then, for each of the 100 sequences, an amino acid was chosen at random and changed to a different amino acid that was also chosen at random. trRosetta then ran a new analysis, and the results were compared; any change that made things look more structured was retained.
By about 20,000 repeats of this process, the compactness of the arrangement of the amino acids in these hallucinations were similar in nature to those of regular proteins. But, critically, the amino acid sequences didn’t look like those of known proteins. The structures themselves didn’t either. In the proteins used by life, there are often loops of poorly structured amino acids that perform key functions. But the hallucinations weren’t selected for function; they were selected for compactness. So, those sorts of extended loops were not found in the hallucinations.
There are a couple of reasons to be skeptical that actual chains of amino acids would form these structures in the real world. trRosetta isn’t the latest and greatest in structure-prediction software that’s been making all the headlines. And trRosetta was trained to figure out structure in part by evaluating evolutionary relationships. These proteins are all brand new and have no evolutionary relatives. The process would only work if the neural network used in trRosetta had inferred principles of protein structure from those evolutionary relationships.
The only way to tell whether it worked is to make the actual proteins and see what they look like. So, the research team put together genes that encoded 129 hallucinatory proteins.