How, exactly, does the coronavirus hijack and reprogram human cells to sicken and kill?
This question has obsessed Nevan Krogan since February, when the UCSF virus expert and his colleagues realized, before many did, that things in America would get very bad very fast.
Since then, the question has only grown more urgent, and for the past four months, Krogan and an ever-expanding team of scientific collaborators in San Francisco and around the world have turned their labs upside down, prying out secrets of the virus that might point to a cure.
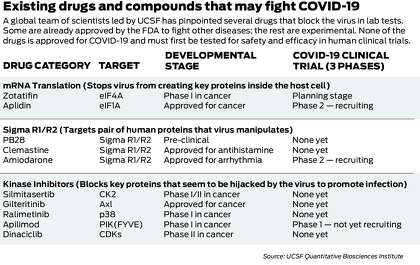
They have built an innovative system for quickly generating clues about the virus’ weak spots and using those clues to search vast drug databases for existing drugs that might stop it in its tracks. First, they assembled a first-of-its-kind map of the virus’ inner workings, exploiting the map to pinpoint 10 old drugs and compounds that kill the virus in lab tests and could ultimately become drugs for treating COVID-19.
Now the UCSF-led group has used similar techniques to explore the biology of the virus at a deeper level, flagging a new class of drug candidates that function through a different, powerful mechanism.
And this time, the scientists have taken photos of the virus as it infects host cells, providing some of the first sharp visual images of the deadly pathogen at work — and exposing weird cellular structures created by the virus that have never been seen before and that may help explain why it is so infectious.
“The more you understand this creature, the better you can fight it,” said Krogan, a molecular biologist at UCSF and an investigator with the Gladstone Institutes who led the large research team. “So we’re trying to understand as much as possible about how the virus infects us.”
Krogan directs the Quantitative Biosciences Institute within UCSF’s School of Pharmacy, a coalition of 100 research laboratories that often work together on projects, generating reams of biological data and sifting those data sets for clues about fighting disease.
The new findings, expected to be published Saturday in the prestigious journal Cell, emerged from a joint effort of 22 QBI labs that are aiming their energies at the virus. Dubbed the QBI COVID-19 Research Group, or QCRG, it brought aboard some 80 scientists in four countries to pursue the project, linking UCSF with teams at the Icahn School of Medicine at Mount Sinai in New York, the Institut Pasteur in Paris, the University of Freiburg in Germany and the European Molecular Biology Laboratory in Cambridge, England.
The discoveries follow innovative research that made a splash starting in March, when the QCRG began releasing a striking set of results published in the journal Nature in late April.
The core of that project was a “protein-protein” map: A comprehensive picture of how the virus takes over human cells by enmeshing its own proteins with human proteins. Viruses can’t survive on their own. They require host cells to reproduce. The map revealed 332 distinct protein-protein interactions — a bounty of intelligence about the virus’ vulnerabilities.
The new research highlighted in Cell goes deeper, building on the map while enlarging the universe of knowledge about the virus, UCSF scientists said.
This time, they explored the virus’ ability to manipulate a powerful biological process called phosphorylation.
During phosphorylation — which is happening all the time inside cells, virus or no virus — certain chemicals get added to proteins or removed from proteins, slightly altering their structure. Because proteins are the cell’s microscopic workhorses, building tissues and sparking the chemical reactions that control all sorts of bodily functions, these deceptively tiny changes can have a large impact on how proteins operate, giving them new abilities. Phosphorylation, then, is like a shortcut for rewiring biology. You don’t have to make a whole new protein to get a job done; you only have to tack on or chop off a little piece of an existing protein.
The tools that do the tacking and chopping are a class of about 500 proteins called kinases, which have powerful effects on human growth, stem cell renewal, the immune system, and memory and learning.
“They are unbelievable at signal processing,” said Kevan Shokat, a co-author of the Cell paper and a UCSF chemist who has studied kinases for 25 years. He described them as a kind of control system, like silicon gates on a computer chip or traffic lights in a city: Kinases appear throughout the cell, regulating the flow of structures and information.
Kinases are also “very druggable,” Krogan said. “A lot of anti-cancer drugs are targeting kinases.”
Scientists don’t have a complete picture of how kinases work. But they know a lot. A major advance was made at UCSF decades ago, earning researchers Michael Bishop and Harold Varmus the 1989 Nobel Prize in medicine. The discovery opened up a new world of drugs that block kinases, known as kinase inhibitors, and since then, about 50 such drugs have been approved by the Food and Drug Administration to fight a range of diseases, mostly cancers, while tens of thousands of others are in the experimental stage.
“It’s amazing what one discovery can catalyze,” Shokat said. “It’s the same thing we want with the virus.”
Starting in March, the UCSF scientists speculated that the coronavirus was exploiting kinases to cause damage in the human body — to grab human proteins and rewire them to do the virus’ bidding.
The researchers believed that if they could learn more about how the virus “speaks” to kinases, they could find drugs to render the virus mute.
Months ago, when the UCSF group created the protein map, they analyzed only one viral protein at a time, using small snippets of the virus instead of the entire virus.
This time, beginning in April, they started by infecting host cells with the live virus. Then, once the infection was raging in the lab plates, the scientists essentially looked over the virus’ shoulder, watching it take over the host cells by analyzing the host proteins as they were altered by the invader.
To pick those proteins out of the mix and get a picture of the virus at work, the team relied on machines called mass spectrometers, which determine the identity and abundance of proteins. A fire hose of protein data soon poured out from the mass spectrometers, and sifting through it all, the scientists started to pick out the specific kinases that the virus seemed to be weaponizing. If a cell is like a city, it was as if the team were floating above the city grid at night and seeing many of the stoplights — the kinases — all at once, blinking green or red.
On several streets, most of the lights were green, meaning that the virus was probably activating those areas to perform jobs that would harm the host. One street that seemed particularly active was ruled by a kinase called casein kinase 2, which happened to be one of the 332 proteins flagged in the team’s earlier protein map.
As clues from the mass spectrometers continued to stack up, the UCSF researchers collaborated with scientists in Germany and Montana to take ultra-close-up snapshots of the virus as it manipulated the host cells, using powerful electron microscopes.
When the team looked at the photos, they got a shock: The surface of the host cell was bristling with wispy, finger-like strands known as filopodia — sharp little straws that aren’t usually there, but had been manufactured by the virus. They were poking out from the inside of the infected cell, cutting holes in the membrane and creating a tunnel to the outside.
Looking closer and performing more tests, the scientists began to get excited because the virus was doing something unexpected.
Normally, the way a virus spreads itself is by turning an infected cell into a virus factory. The cell fills with virus copies like a water balloon, finally bursting and releasing the virus particles. But the filopodia seemed to show that the virus has devised an additional method for promoting infection: After copying itself within the host cell — but before the cell bursts — the virus exits through filopodia that tunnel out from the infected cell and punch a hole in a nearby uninfected cell, allowing the virus to swim from one cell to another.

Similar filopodia have been detected in other viruses, like smallpox, but Krogan said they’ve never been seen to this extent. Tests and electron microscope photos showed that these filopodia were packed with copies of the coronavirus as well as Casein Kinase II. The photos — captured by Elizabeth Fischer, chief of the microscopy unit at Rocky Mountain Laboratories, and scientists at the University of Freiburg — revealed that the virus was “budding” out from the filopodia, and, incredibly, some of the filopodia could even branch like trees, allowing a single strand to punch holes in two cells at once.
“It’s so biologically revealing,” Krogan said of the photos, calling them “the most fascinating and awful thing I’ve ever seen.”
“It’s unbelievable what this virus can do,” he added.
Now that the scientists had a better sense of which kinases were being manipulated by the virus, they pored through databases of existing drugs and experimental compounds to find chemicals that might inhibit those specific kinases and one day become drugs against COVID-19.
They identified 87 kinase inhibitors that could plausibly do the job, including 10 that were already FDA-approved to treat other diseases, 53 being tested in human trials and 24 that were “preclinical,” not yet given to humans. Shokat happens to keep thousands of kinase inhibitors in a freezer at UCSF (Krogan calls it “the biggest freezer in the world” for this type of molecule), and from the full list of 87, they selected 68 candidates to test against the virus in the lab.
The tests were performed by virologists at Mount Sinai and the Institut Pasteur, who infected host cells with the virus and added the kinase inhibitors. Would they kill the virus? Would they flip the cellular traffic lights from green to red, blocking the harmful kinases?
In the end, more than a dozen of the 68 candidates looked promising. The team narrowed down the list to seven or eight that were particularly potent — including one that blocked Casein Kinase II. And five showed greater killing power in the lab setting than remdesivir, the only antiviral drug currently available to some patients for emergency use (in addition, a steroid called dexamethasone has been found to reduce the COVID-19 death rate).
There are still things the scientists don’t know about how the coronavirus infects cells. They want to understand why the virus selects and exploits certain kinases; they also aren’t sure if the filopodia stop forming when Casein Kinase II is blocked. As for the drug candidates flagged in the new paper, they must be tested in human trials to see if they are safe and effective for COVID-19 patients.
But Krogan said he is optimistic that some will pan out. According to a recent tally by Nature Biotechnology, the research efforts led by UCSF have spawned more than a dozen clinical trials of potential COVID-19 therapies. Ultimately, if the trials go well, some of these drugs could be integrated with remdesivir to create a cocktail therapy for COVID-19.
“I have zero doubt that there’s a cocktail out there that can cure this,” Krogan said. “It’s just: Can we be smart enough, quick enough, to find it?”
Jason Fagone is a San Francisco Chronicle staff writer. Email: jason.fagone@sfchronicle.com Twitter: @jfagone